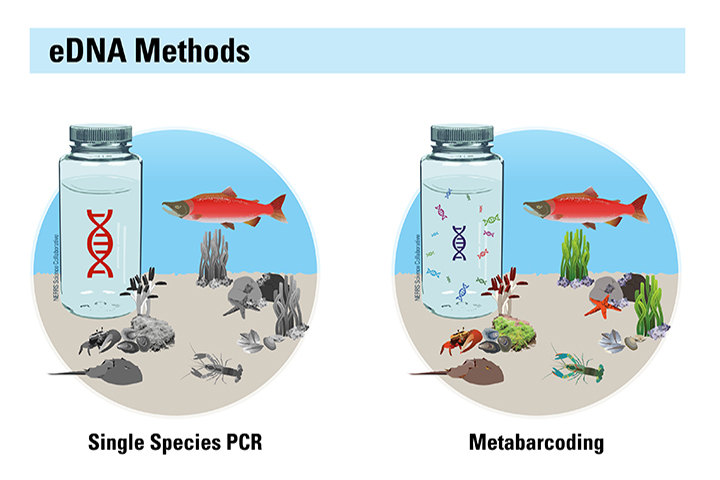
Biological monitoring programs are essential foundations for effective management of estuaries and coasts, but they can be expensive to conduct and methods such as the capture of living organisms may be traumatic for some target species. Advancements in DNA methods now make it possible to identify the organisms in an area by the DNA they leave behind. Environmental DNA (eDNA) comes from feces, gametes, scales, and cells that an organism sheds, and is easily collected from water and sediment samples. Rapid reductions in analytical costs now allow scientists to analyze eDNA in water samples and identify dozens of species without having to capture live animals or plants; however, when this project began there were few proven methods suitable for highly variable estuarine conditions.
This project established a Community of Practice around eDNA sampling within the National Estuarine Research Reserve system and explored the potential for eDNA to support estuarine management. Scientists and staff from six reserves worked collaboratively with researchers, resource agencies, and a technical advisory team to design and implement eDNA-based monitoring protocols for specific issues and species of interest in estuarine ecosystems. The project found that fish community and biodiversity assessments are well suited to eDNA applications, while invasive crabs are much harder to detect because they do not shed much DNA. Researchers tested several methods to collect, filter, and extract DNA and developed protocols and how-to guides for practical, time efficient approaches that work in unique estuarine conditions. Several reserve sites are now integrating eDNA into their monitoring programs to track fish species that can be efficiently monitored with eDNA.